|
Introduction
The quantitative
analysis of confocal microscopy data has several phases from data generation
to statistical analysis. Broadly we regard the following topics:
-
image extraction
-
image analysis and boundary
detection
-
extraction of point patterns for
foci of interest (nuclear compartments, genomic loci)
-
analysis of colocalization in the
point patterns
-
formulation of descriptive or
discriminative models for cell speciation
-
formulation of generative
mathematical/probabilistic models for system development.
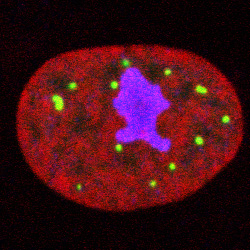
Nucleus Image: PML foci (green) distributed in
cell nucleus (red)
with nucleolus (purple) Here we focus on the
analysis of point pattern data, in the absence of information concerning the
location of the nuclear boundary, or exclusion regions such as
nucleoli.
We refer to the compartments or domains as object types, and the
individual foci as objects.
We aim to assess the following two (null)
hypotheses:
-
The observed arrangement of foci
corresponding to a single object type is random in the spatial region
available (that is, the cell nucleus, possibly with identified exclusion
regions).
-
The observed arrangement of foci
corresponding to a multiple object types (say, types considered
pairwise, or in triples) does not indicate any spatial
association or dependence between the object types.
Rejection of (A) allows us to
conclude that there is an underlying organizational scheme active in the
compartments of interphase nuclei. Rejection of (B) allows us to
propose that there is a functional association between different object
types; functional interaction is necessarily associated with physical
proximity between the interacting objects.
On to page 2:
Pages 1
2 3
4 5
6 7
Notes:
- These pages
are an attempt to make the statistical analysis of 3D point pattern data
accessible to biologists; statistical jargon and mathematical notation is
therefore kept to a minimum.
- I might fail with 1. ... so
if you have any comments or suggestions, please email
me
- Pages under constant development
Back to Top Back to the BioSPP Home Page |
