
Imperial College London
Department of Mathematics and Division of Molecular Biosciences
 |
Imperial College London Department of Mathematics and Division of Molecular Biosciences
|
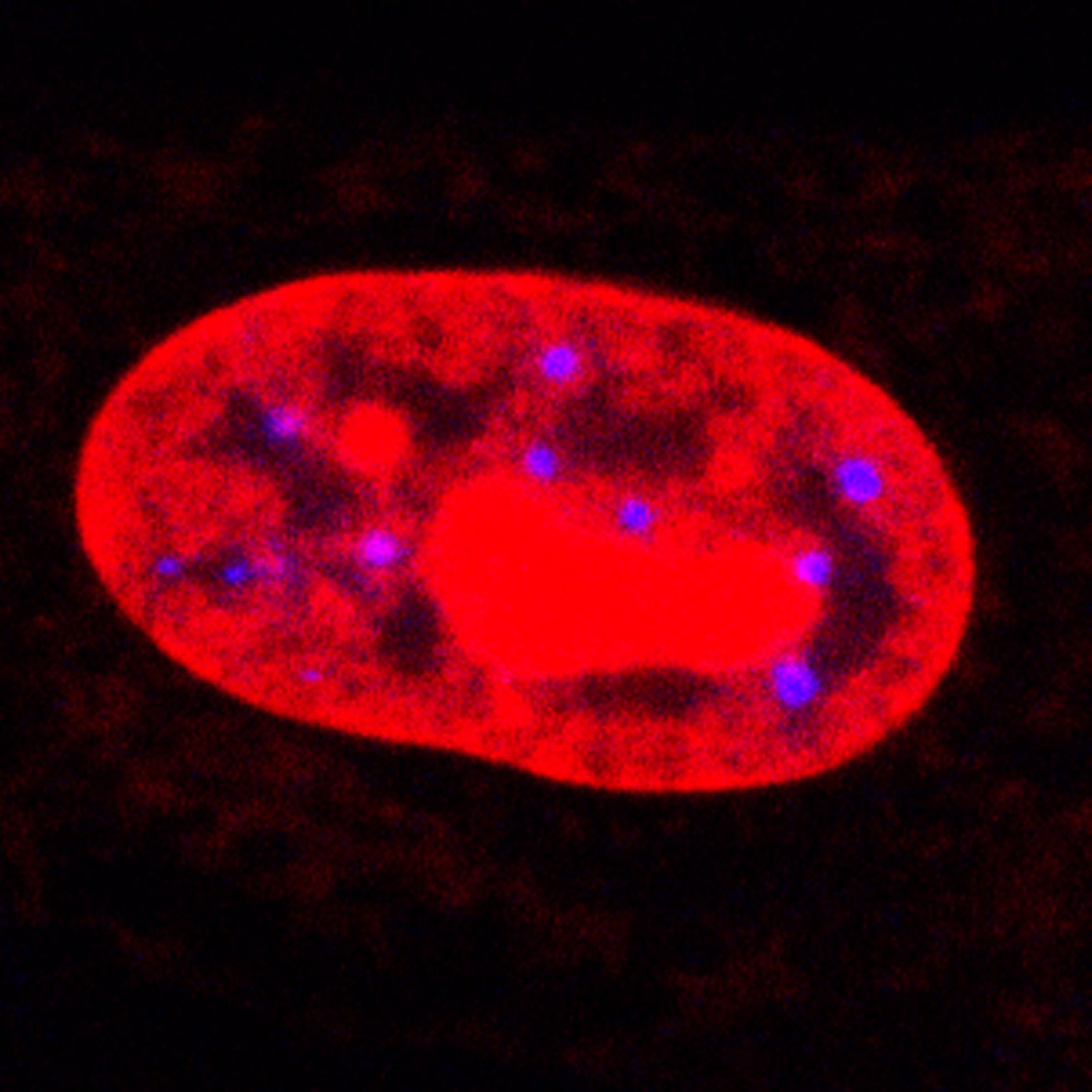
Confocal microscopic image of Promyelocytic Leukemia (PML) in a human cell nucleus.
PMLs stained blue.
|
These pages describe work carried out at Imperial College on the statistical analysis of spatial point patterns (SPPs) that arise in the imaging of mammalian cell nuclei. The observed point patterns correspond to the locations of nuclear domains collected using confocal microscopy. The specific biological objective is to understand positional and functional nuclear organization, that is, how the different domains and compartments colocalize spatially, as it is believed that this gives an insight into biological function and interaction. We have specifically focussed on the investigation of the Promyelocytic Leukemia (PML) body nuclear domain (see also here), and the related domain CREB Binding Protein (CBP) foci, and their spatial relation to genomic and other loci. Different aspects of the analysis problem are described in the following sections: |
||||||||||||||||||||||||||||||
|
People involved in the work: Faculty
PhD Students
Collaborators
|
||||||||||||||||||||||||||||||
Some PML Images
Computation
We recommend computation using the statistical package R
Some code in R for BioSPP analysis
Other Material
Funding: Studentships provided by UK Research councils EPSRC, MRC, BBSRC
This page created by David Stephens; all content © Imperial College 2006.