research
stochastic reaction networks
Cells proliferate by constantly producing molecules that carry out essential functions. Many of these molecules are present in such low numbers that reaction rates fluctuate from cell to cell and over time. We develop stochastic methods based on the Chemical Master Equation to quantify how cells can function reliably despite this stochasticity and how they diversify in response to external stimuli.
highlights:
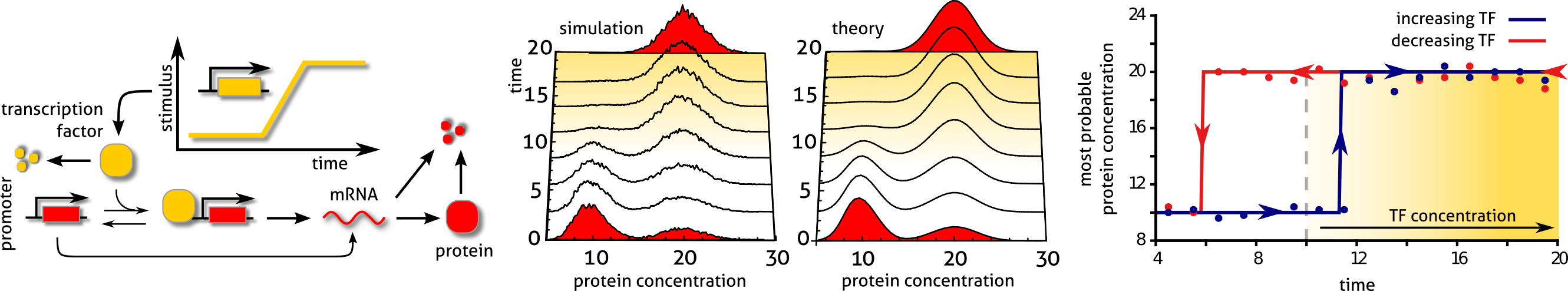 Noise-induced bimodality and hysteresis in a phenotypic switch encoded by a gene expression network. PNAS 111:6994
Noise-induced bimodality and hysteresis in a phenotypic switch encoded by a gene expression network. PNAS 111:6994
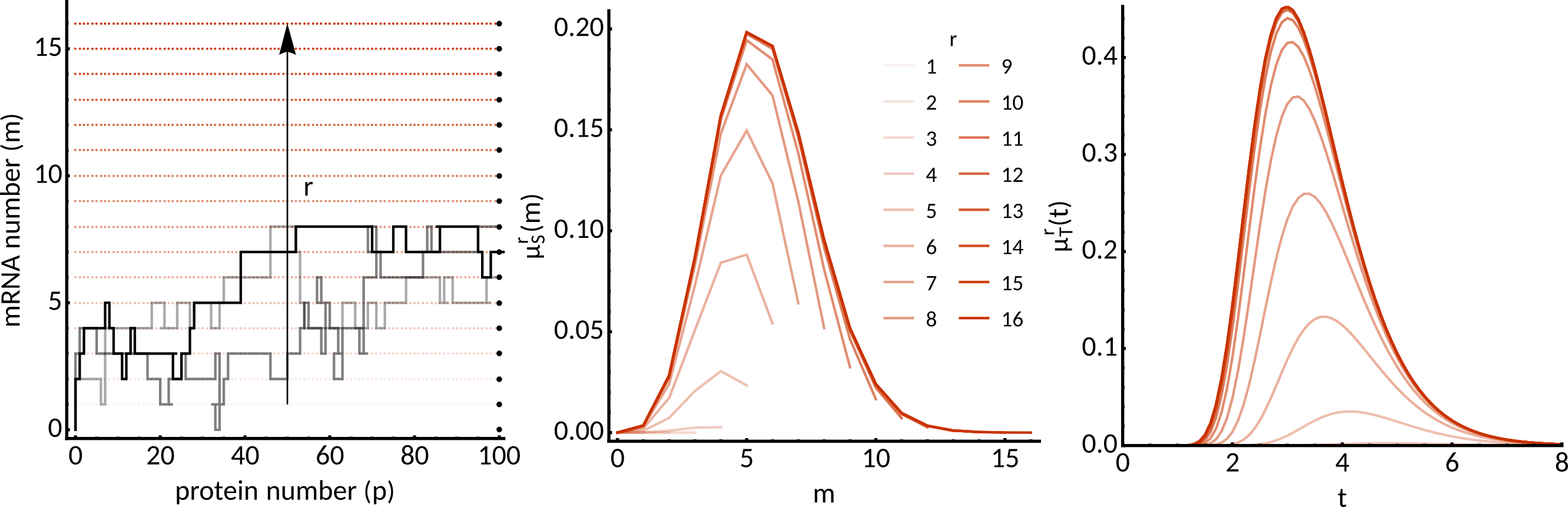 Exit-time finite state projection algorithm: computation of first-passage-time and exit location of a protein threshold. SIAM Sci Comput 41, A748-A769
Exit-time finite state projection algorithm: computation of first-passage-time and exit location of a protein threshold. SIAM Sci Comput 41, A748-A769
agent-based models of cell populations
Cell divisions generate ancestral relationships between cells in a population, which are called lineage trees, and can be observed under a microscope. Disentangling these genealogical relationships allows us to make sense of cell fate decision and cellular heterogeneity. To do this, we develop mathematical methods to predict cellular histories from agent-based models of growing and dividing cell populations.
highlights:
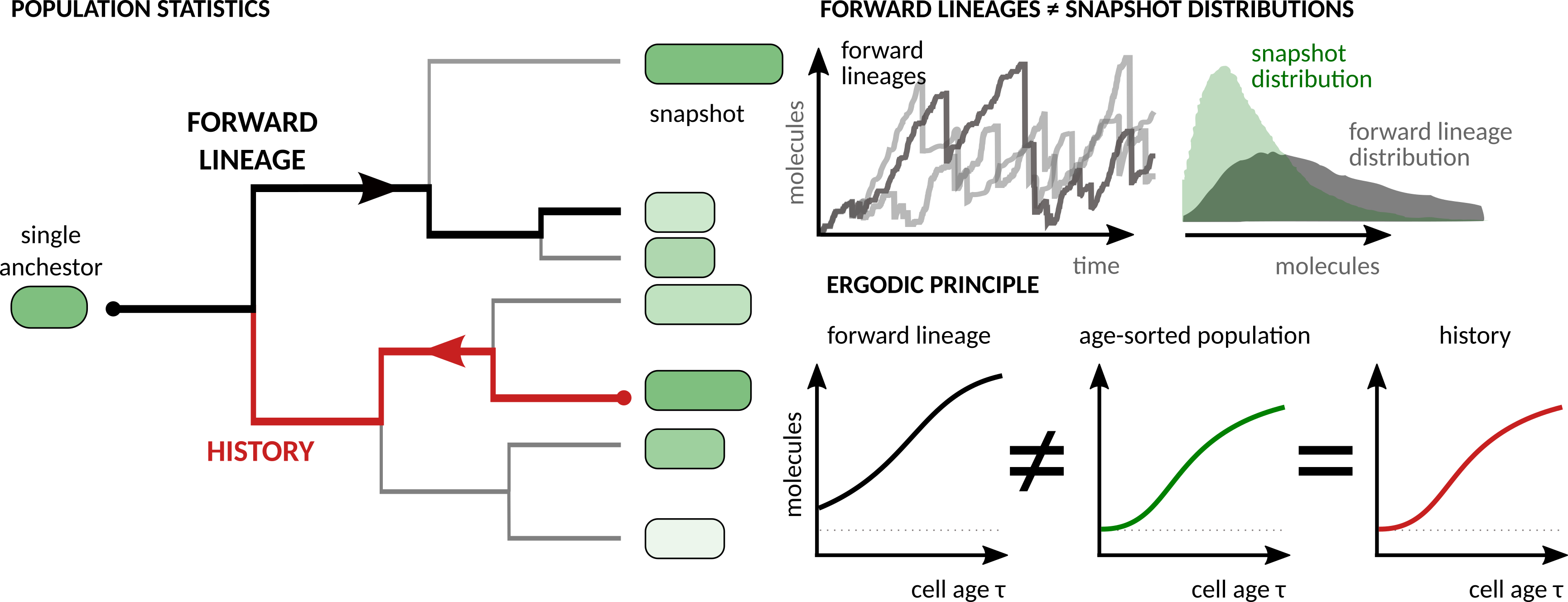 Ergodic principle relating the cell-to-cell heterogeneity of gene expression in a population of dividing cellular agents to their cell histories. J Royal Soc Interface 14, 20170467
Ergodic principle relating the cell-to-cell heterogeneity of gene expression in a population of dividing cellular agents to their cell histories. J Royal Soc Interface 14, 20170467
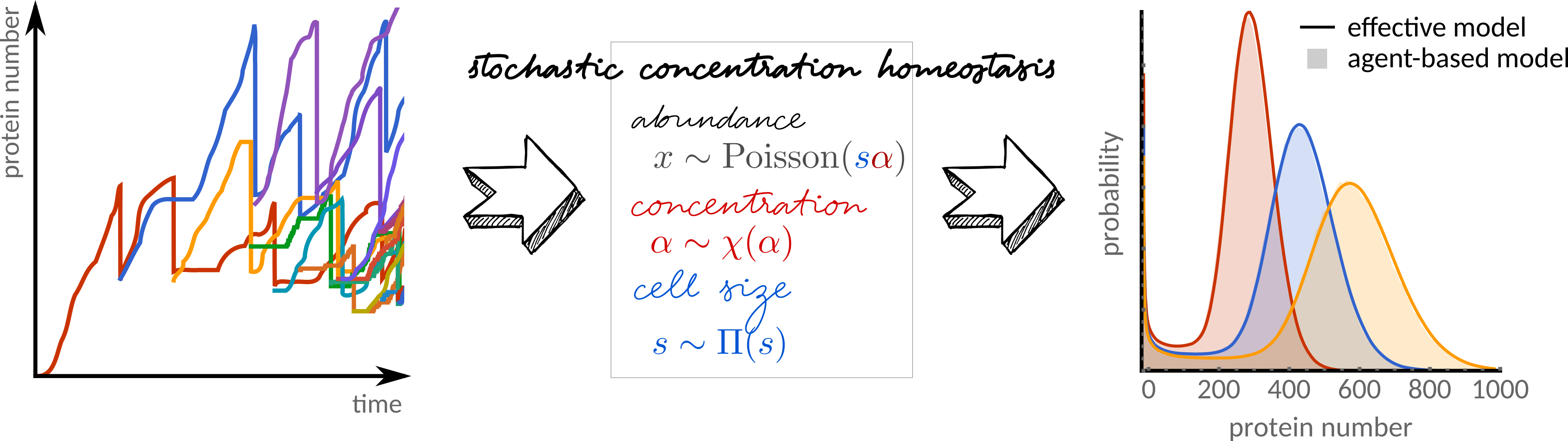 Stochastic concentration homeostasis guarantees the validity of effective dilution and extrinsic noise models of cell-size-dependent gene regulatory networks. J Royal Soc Interface 18, 20210274
Stochastic concentration homeostasis guarantees the validity of effective dilution and extrinsic noise models of cell-size-dependent gene regulatory networks. J Royal Soc Interface 18, 20210274
 Cell cycle phase inheritance models to reveal biological oscillators that drive the cell cycle. (In preparation)
Cell cycle phase inheritance models to reveal biological oscillators that drive the cell cycle. (In preparation)
cell models
Cells constantly make decisions such as when and at what size to divide. These decision processes may appear random without investigating which internal and external signals cells may respond to. We develop stochastic models that describe how and why cells make decisions. Using these models we infer which molecular networks are at play and identify environmental factors affecting these decisions.
highlights:
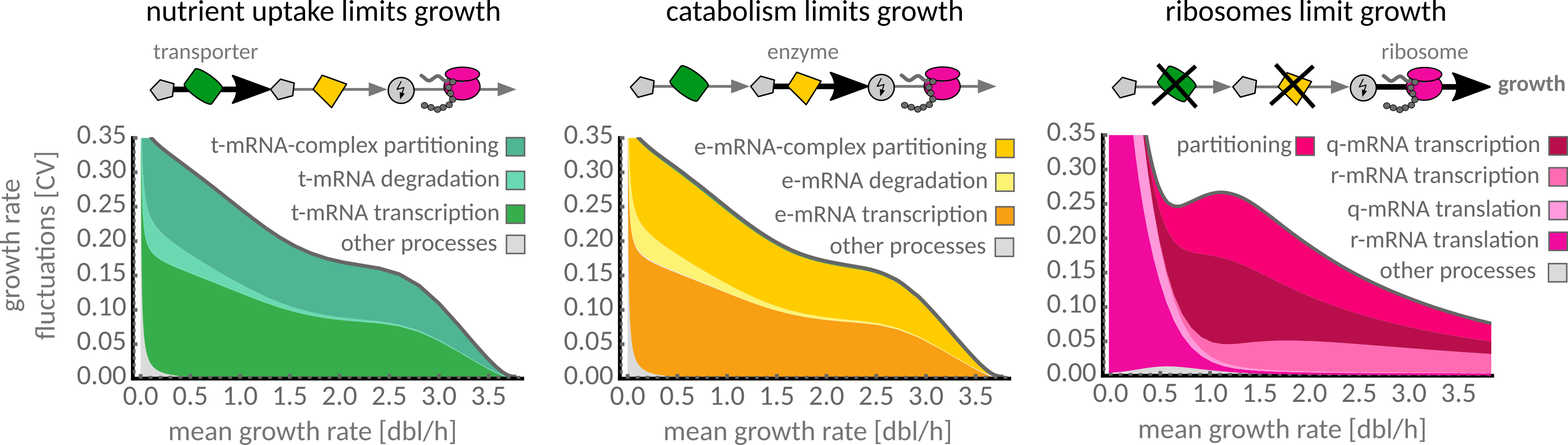 Coarse-grained cell model predicts the propagation of gene expression noise to growth rate fluctuations under different growth limitations. Nat Commun 9, 4528
Coarse-grained cell model predicts the propagation of gene expression noise to growth rate fluctuations under different growth limitations. Nat Commun 9, 4528
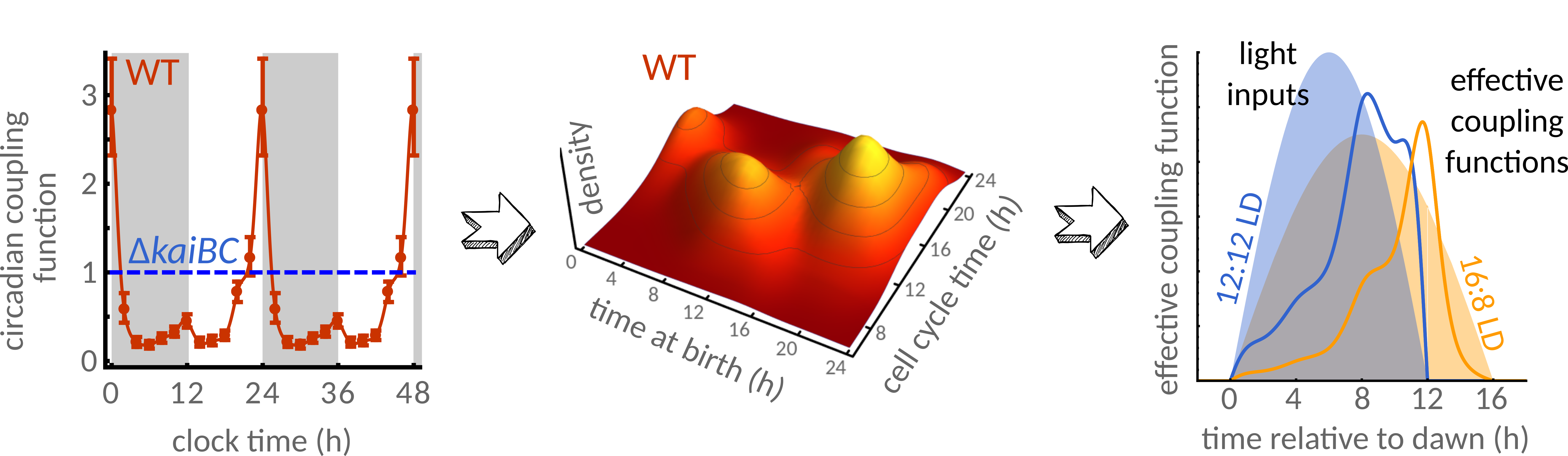 Coupling of cell divisions to circadian rhythms allows us to infer the response of cells under time-varying environmental conditions. PNAS 115, E11415-E11424
Coupling of cell divisions to circadian rhythms allows us to infer the response of cells under time-varying environmental conditions. PNAS 115, E11415-E11424
collaborators
-
Dr Alexis Barr (LMS-MRC, ICL): Cell cycle control in cancer
-
Dr James Locke (Sainsbury Lab, U Cambridge): Circadian clock dynamics in single cells
-
Prof Luca Magnani (Medicine, ICL): Single-cell analysis of dormancy in breast cancer
-
Dr Bruno Martins (Life Sciences, U Warwick): Cell size control in cyanobacteria
in the news
-
Our group received generous funding through a UKRI Future Leaders Fellowship 2020 to advance agent-based modelling approaches for single-cell dynamics.
-
A feature on our work on single-cell growth and its implications for antibiotic tolerance.
-
Two recent news articles (and here) describe our findings about how cells coordinate divisions with their internal clocks and the environment.
 Mycobacterial cell size control in different growth media.
Mycobacterial cell size control in different growth media.